Getting Started with Bioinformatics on Tufts HPC
Shirley Li, Bioinformatician
Yucheng Zhang, Senior Bioinformatics Engineer
Tufts Technology Services, Research Technology
October 2, 2025
Welcome
This workshop is designed to help you get started with bioinformatics on the Tufts High-Performance Computing (HPC) cluster. We will cover:
- Introduction to the Linux/Unix Command Line
- Bioinformatics on the Tufts HPC: tools, workflows, and available resources
Prerequisites
You will need an active Tufts HPC account. If you are new to the HPC, we strongly recommend reviewing our recent Intro to HPC workshop.
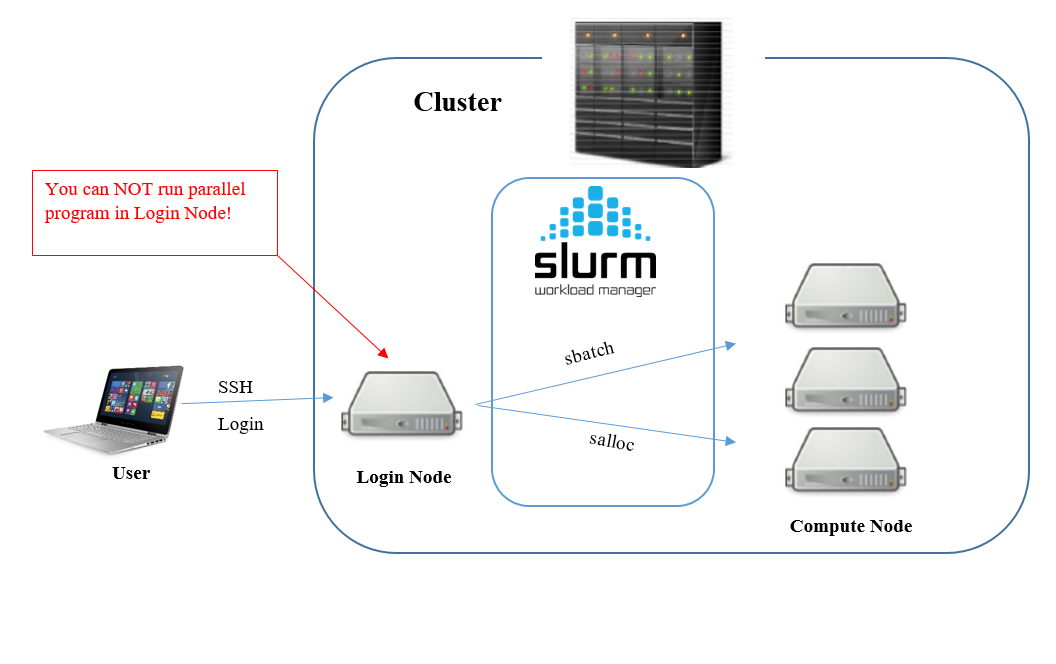
Connecting to the Cluster & Starting a Job
You can connect to the cluster either through a terminal application (such as Terminal on macOS or MobaXterm on Windows) or via the web-based Open OnDemand portal.
Using the Command Line (SSH)
To connect, use the ssh command with your Tufts UTLN.
- New Cluster:
ssh your_utln@login-prod.pax.tufts.edu - Old Cluster:
ssh your_utln@login.pax.tufts.edu
To start an interactive session on a compute node, use the srun command:
srun -N1 -n2 -t2:00:00 --pty bash
This command requests an interactive session with the following resources:
-N1: 1 node-n2: 2 CPU cores-t2:00:00: A time limit of 2 hours
Using Open OnDemand
You can also access the cluster through a web browser via Open OnDemand.
- New Open OnDemand: https://ondemand-prod.pax.tufts.edu
- Old Open OnDemand: https://ondemand.pax.tufts.edu
Recording
Getting started with bioinformatics - 2025 Fall
Upcoming Research Technology Workshops
- Introduction to Programming with Python
- October 9, 2025, 3:00pm - 5:00pm
- Introduction to Git & GitHub
- October 15, 2025, 1:00pm - 3:00pm
- Introduction to Research Computing Tiered Storage
- Tuesday, October 21, 2025, 11:00am - 12:00pm
- Using the newly upgraded HPC Cluster
- Thursday, October 23, 2025, 1:00pm - 3:00pm
- Best Practices for using GPUs in Jupyter for Data Science
- Wednesday, October 29, 2025, 2:00pm - 3:30pm
For a full list, please visit: https://go.tufts.edu/workshops