Pulling, running and building containerized HPC applications
singularity pull
Syntax
Download or pull a container from a given URI.
$ singularity pull [output file] <URI>
Load modules
Like I mentioned in the presentation, to pull or run containers, singularity instead of apptainer is recommended.
$ module purge
$ module load singularity/3.8.4
$ module list
Currently Loaded Modulefiles:
1) squashfs/4.4 2) singularity/3.8.4(default)
Blast
BioContainers is integrated with bioconda. You can find almost all bioinformatics applications from BioContainer’s Package Index
Blast is a widely used bioinformatics application. Here I will show how to pull its latest version (2.15.0) from BioContainers.
Here is the Blast’s bioconda page.
The following is the pull command as listed in the instructions:
docker pull quay.io/biocontainers/blast:<tag>
(see `blast/tags`_ for valid values for ``<tag>``)
You can find the tags by clicking on the container, as illustrated below.

There are different tags for each version and multiple containers/tags even exist for the same application version. It is common practice to select the ones that were last modified.
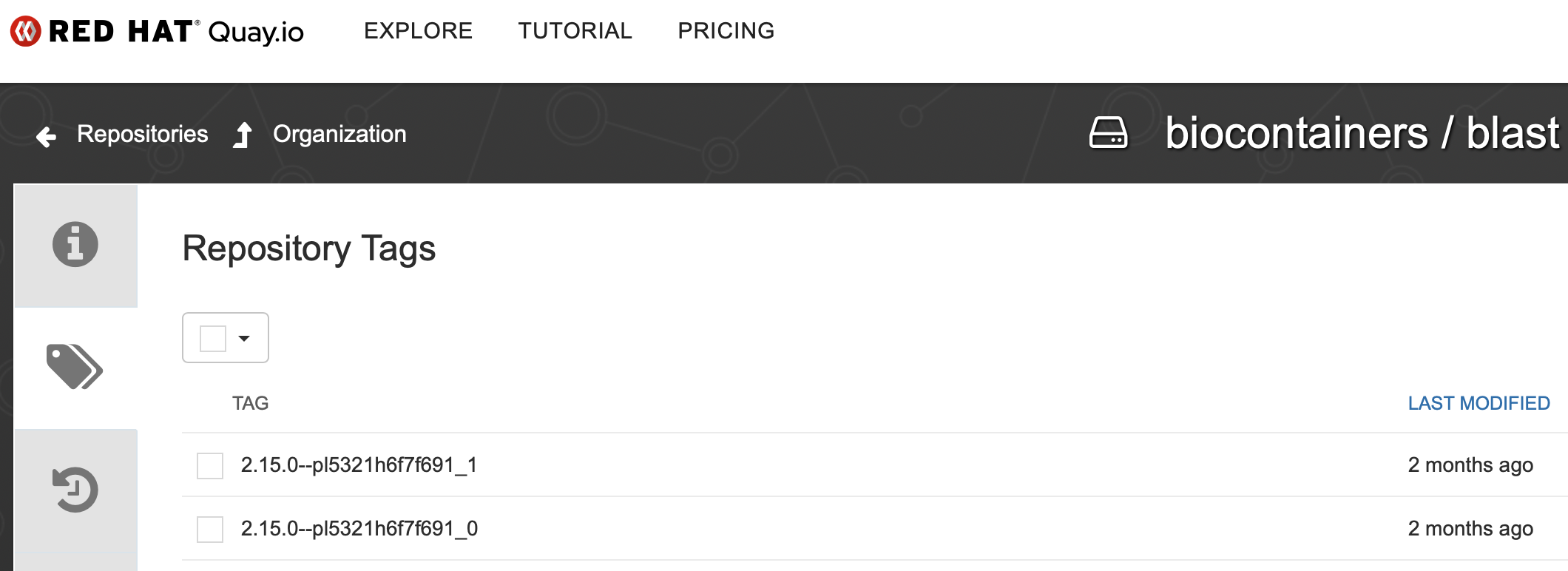
To pull the image from BioContainers, we can use the following command:
## Default
$ singularity pull docker://quay.io/biocontainers/blast:2.15.0--pl5321h6f7f691_1
## To give a customerized output name
$ singularity pull blast_2.15.0.sif docker://quay.io/biocontainers/blast:2.15.0--pl5321h6f7f691_1
$ ls
blast_2.15.0.sif blast_2.15.0--pl5321h6f7f691_1.sif
You will notice that pulling blast_2.15.0.sif is much faster because it utilizes the singularity caches stored in $HOME/.singularity.
PyTorch
PyTorch is a powerful open-source machine learning framework based on the Python programming language and the Torch library. It’s widely used for deep learning, a type of machine learning that builds complex models like artificial neural networks for tasks like image recognition, natural language processing, and more.
If you want to install PyTorch, it can be complex. You have to ensure different libraries (e.g., CUDA) and dependencies are compatible. If you use container, you can easily pull official container image from Docker Hub.
$ singularity pull docker://pytorch/pytorch:2.1.2-cuda11.8-cudnn8-runtime
$ ls
blast_2.15.0.sif* blast_2.15.0--pl5321h6f7f691_1.sif* pytorch_2.1.2-cuda11.8-cudnn8-runtime.sif*
Because pytorch image is large, pulling will take some time. Since we do not have time in the workshop, you can just just copy my pre-pulled image to your current folder and use it for the tests in next sessions.
cp /cluster/tufts/biocontainers/workshop/Spring2024Container/pytorch_2.1.2-cuda11.8-cudnn8-runtime.sif .